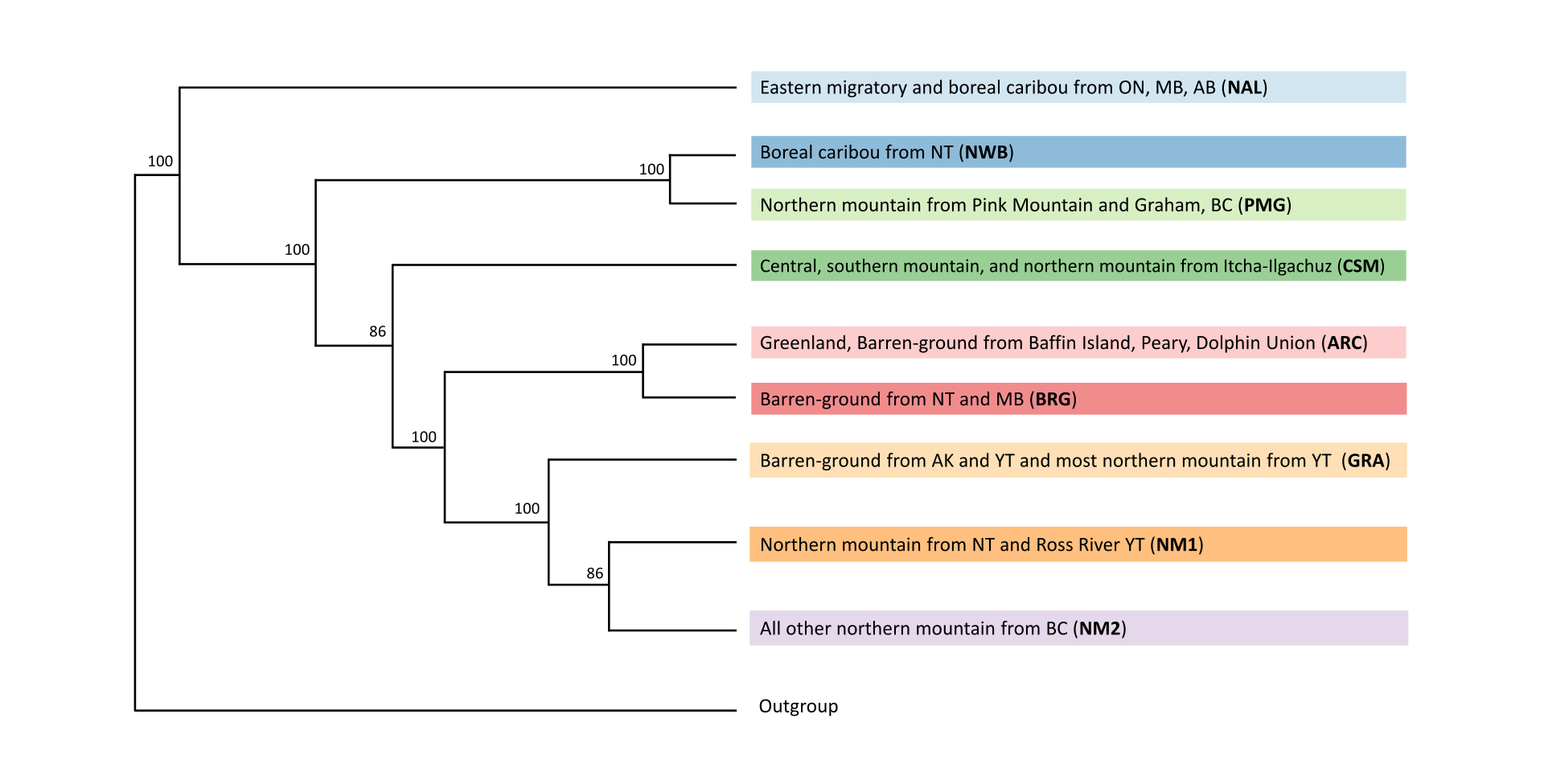
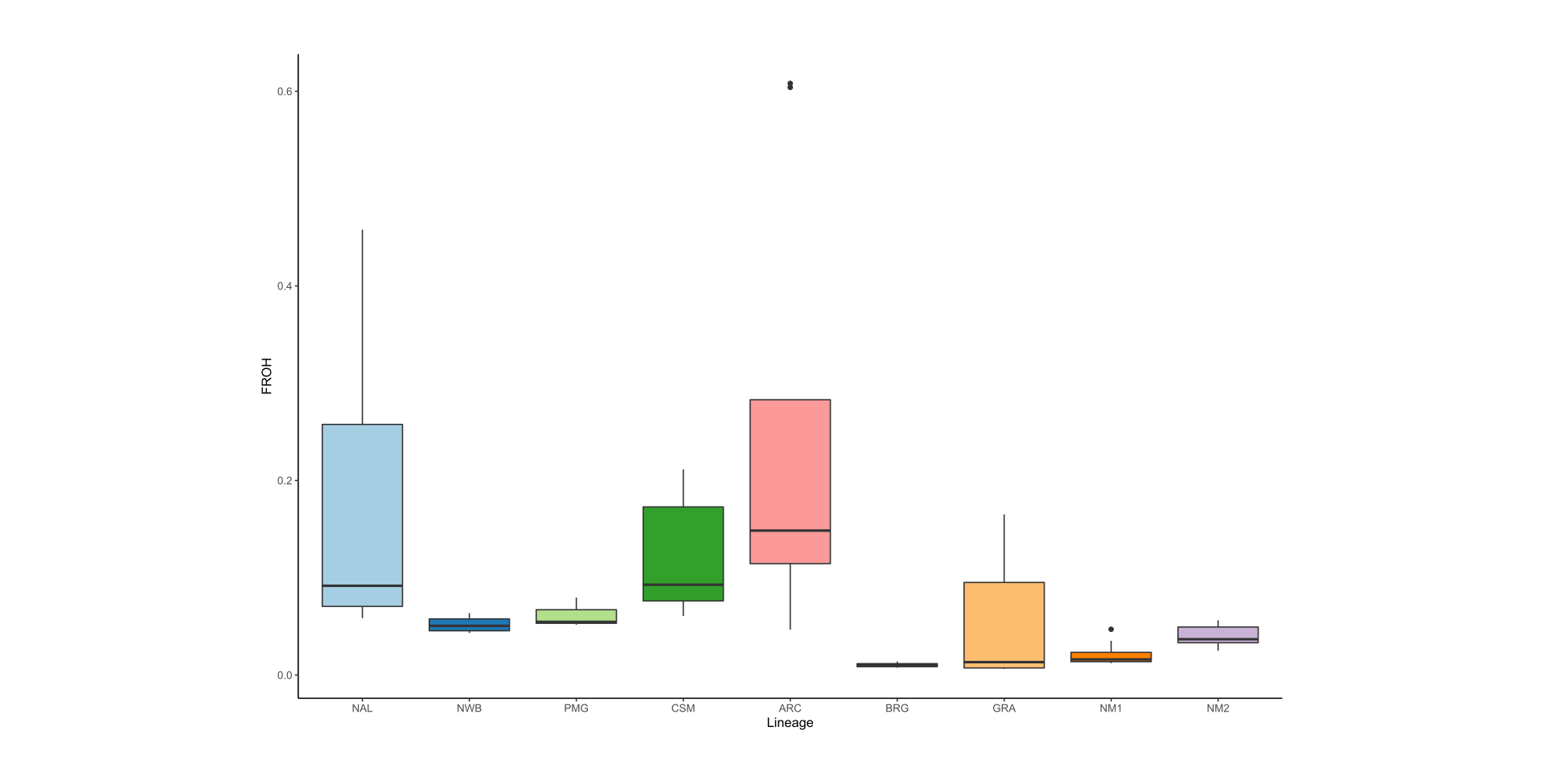
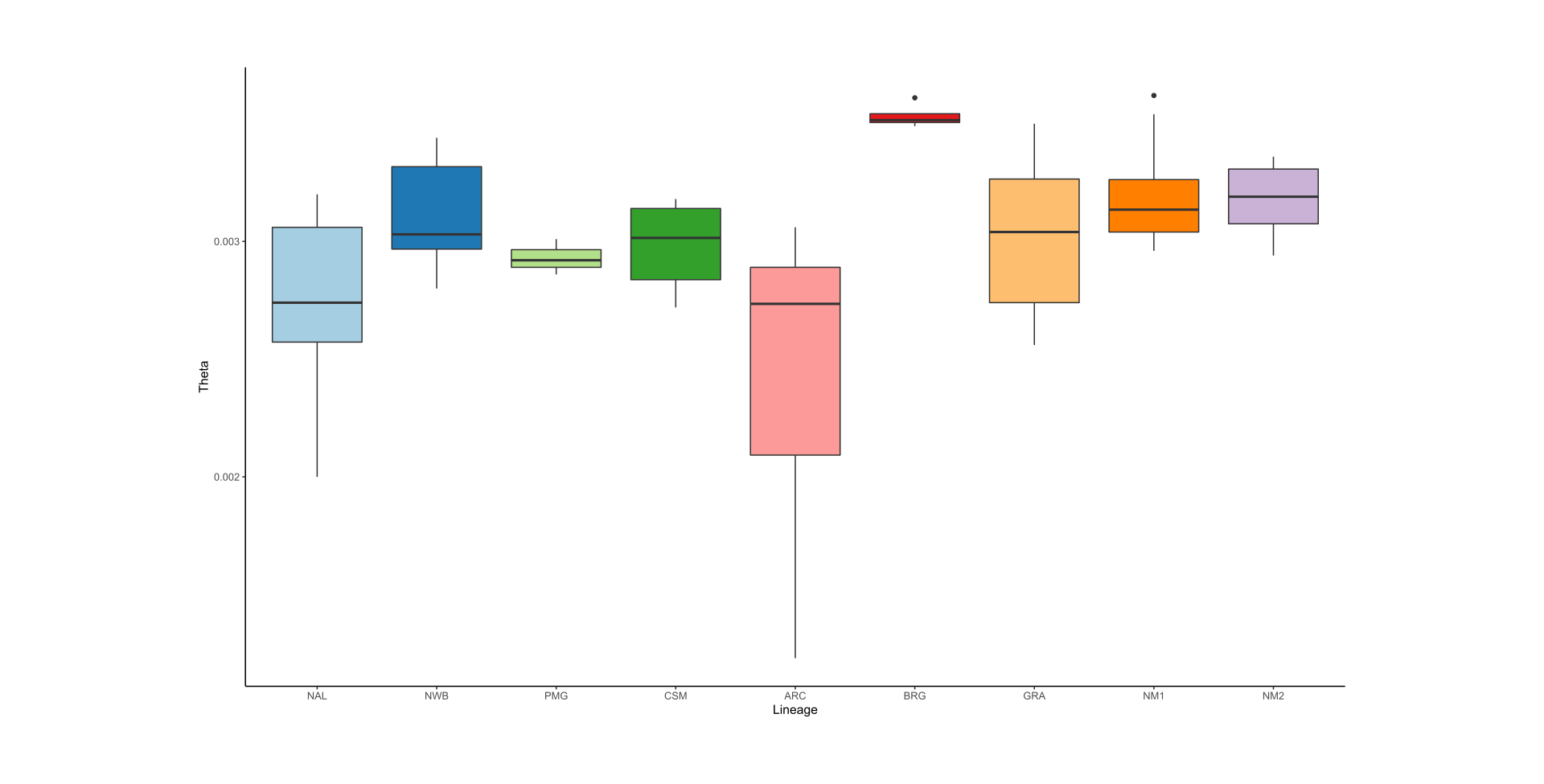
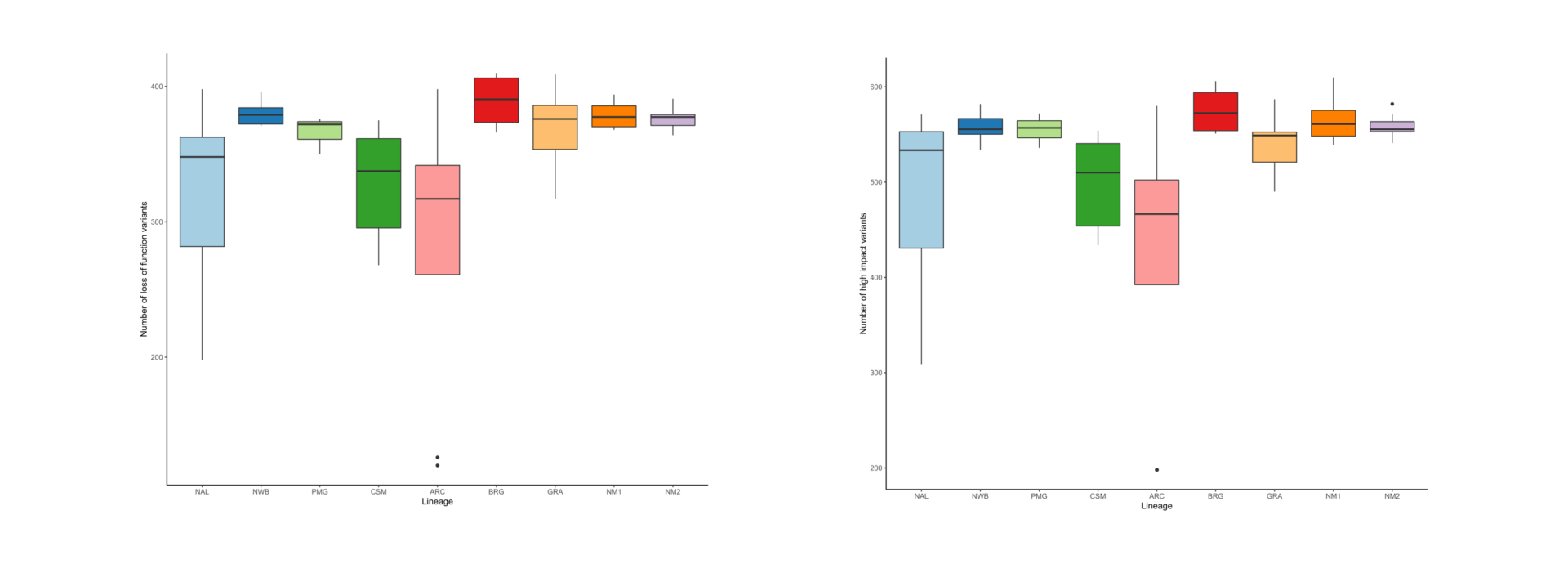
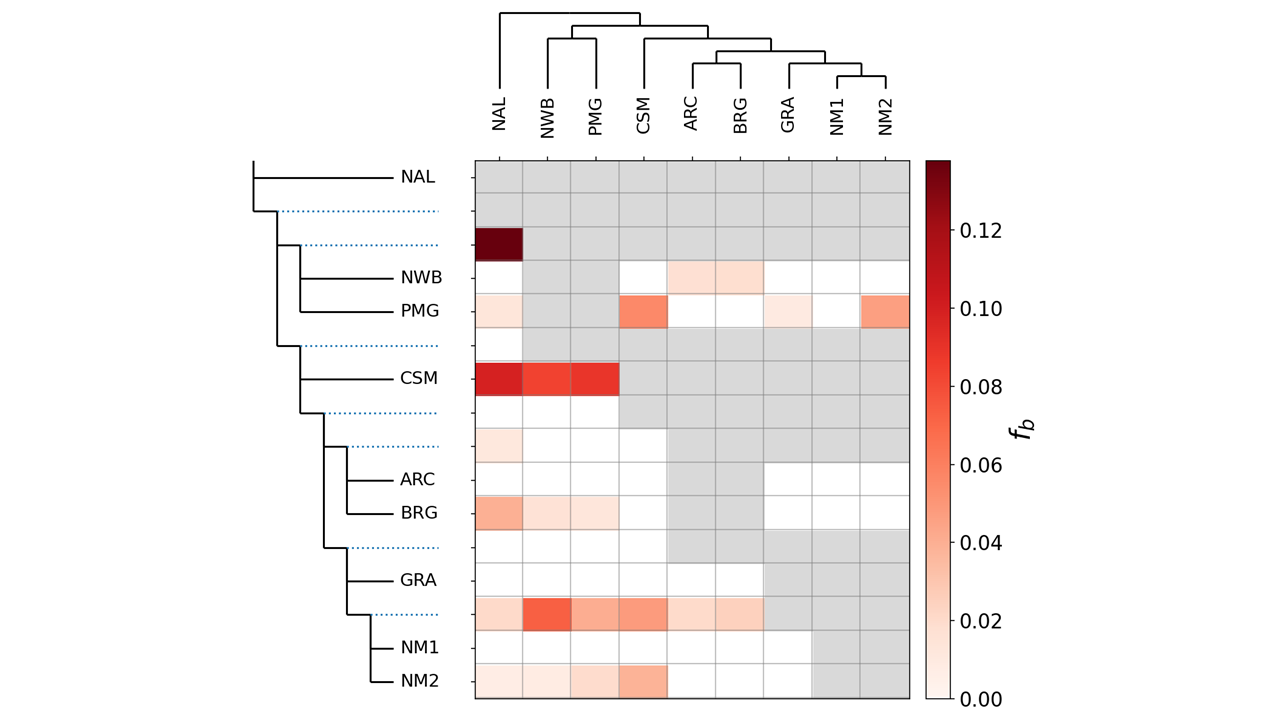
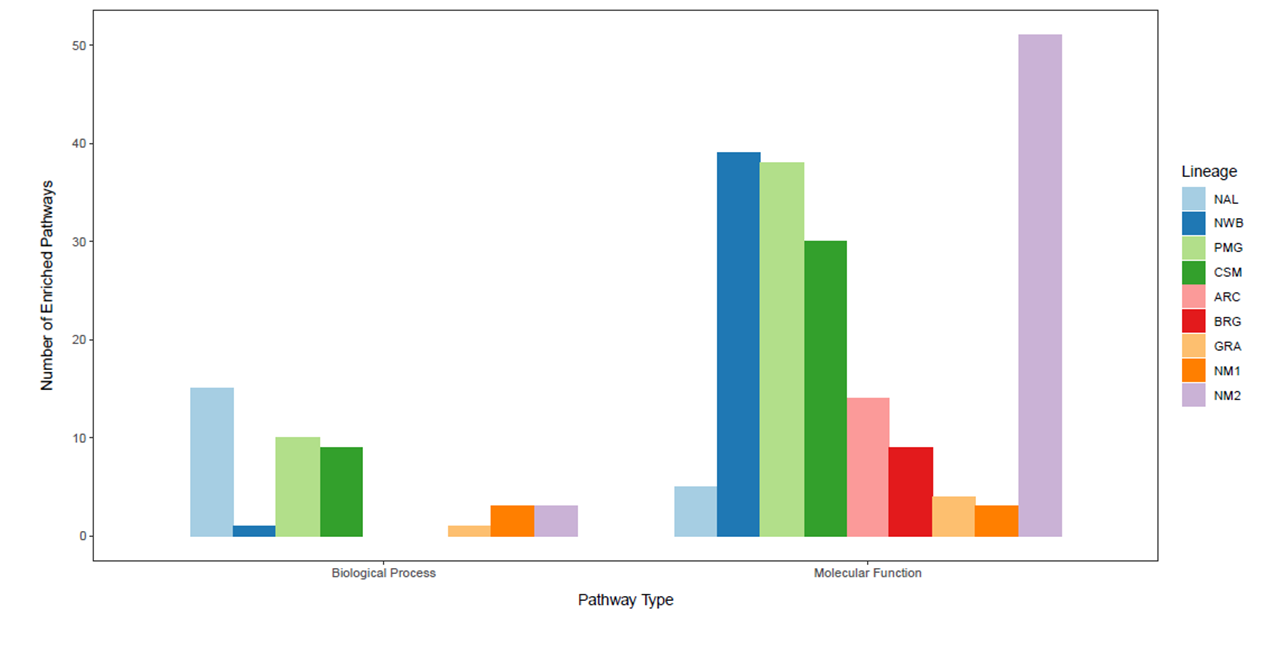
As part of the research program of the Ecogenomics team, we are sequencing and analyzing whole genome sequences of caribou (Rangifer tarandus) to answer questions pertinent to the conservation of the species. So far, we have generated a chromosome scale reference genome and high-quality annotation with Dovetail Genomics, as well as both high coverage (~100) and low coverage (~350) re-sequenced genomes.
Here is an overview of the whole genome sequencing work completed thus far, also see the publications and resources pages for more information:
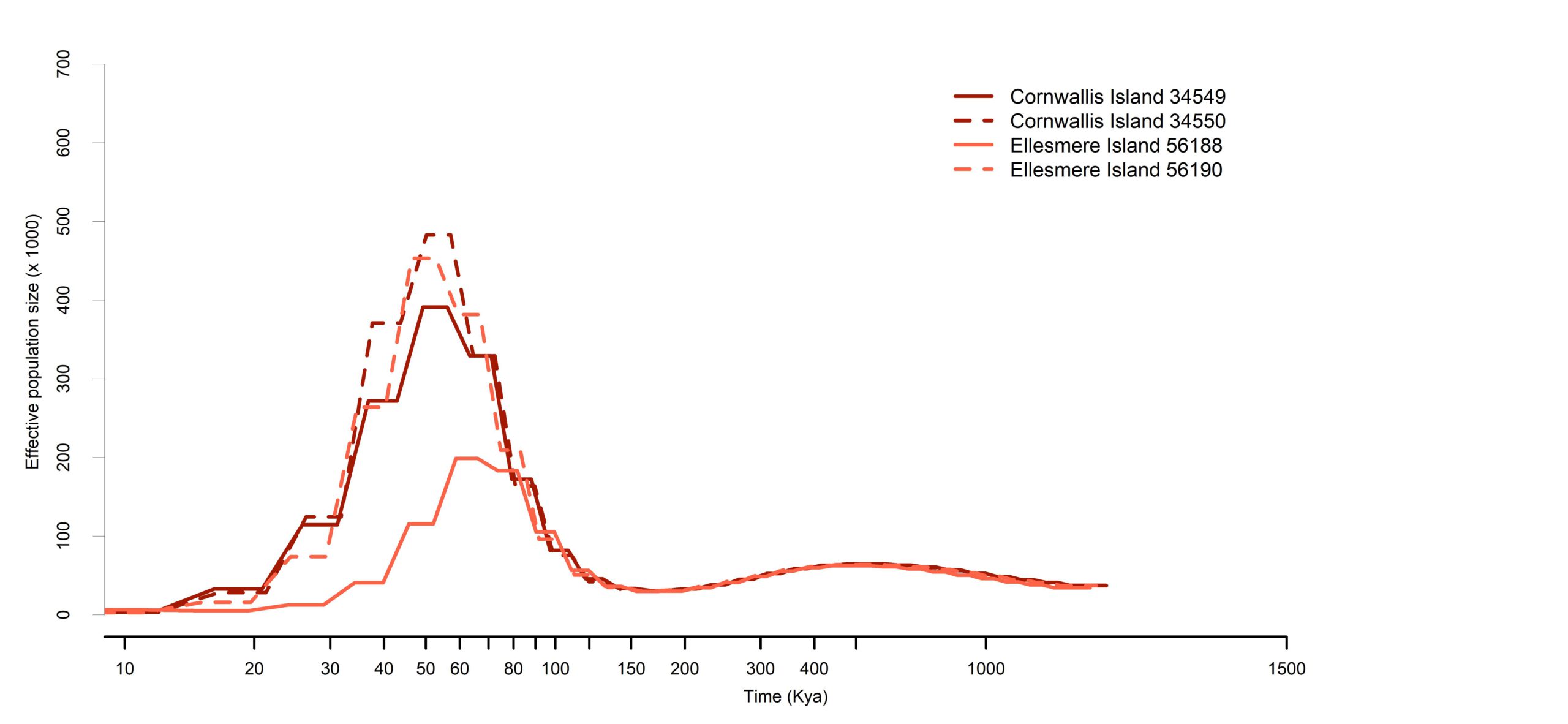
We reconstruct historical Ne to help understand demographic history, responses to past climate changes, and the likelihood of having high genetic variation.
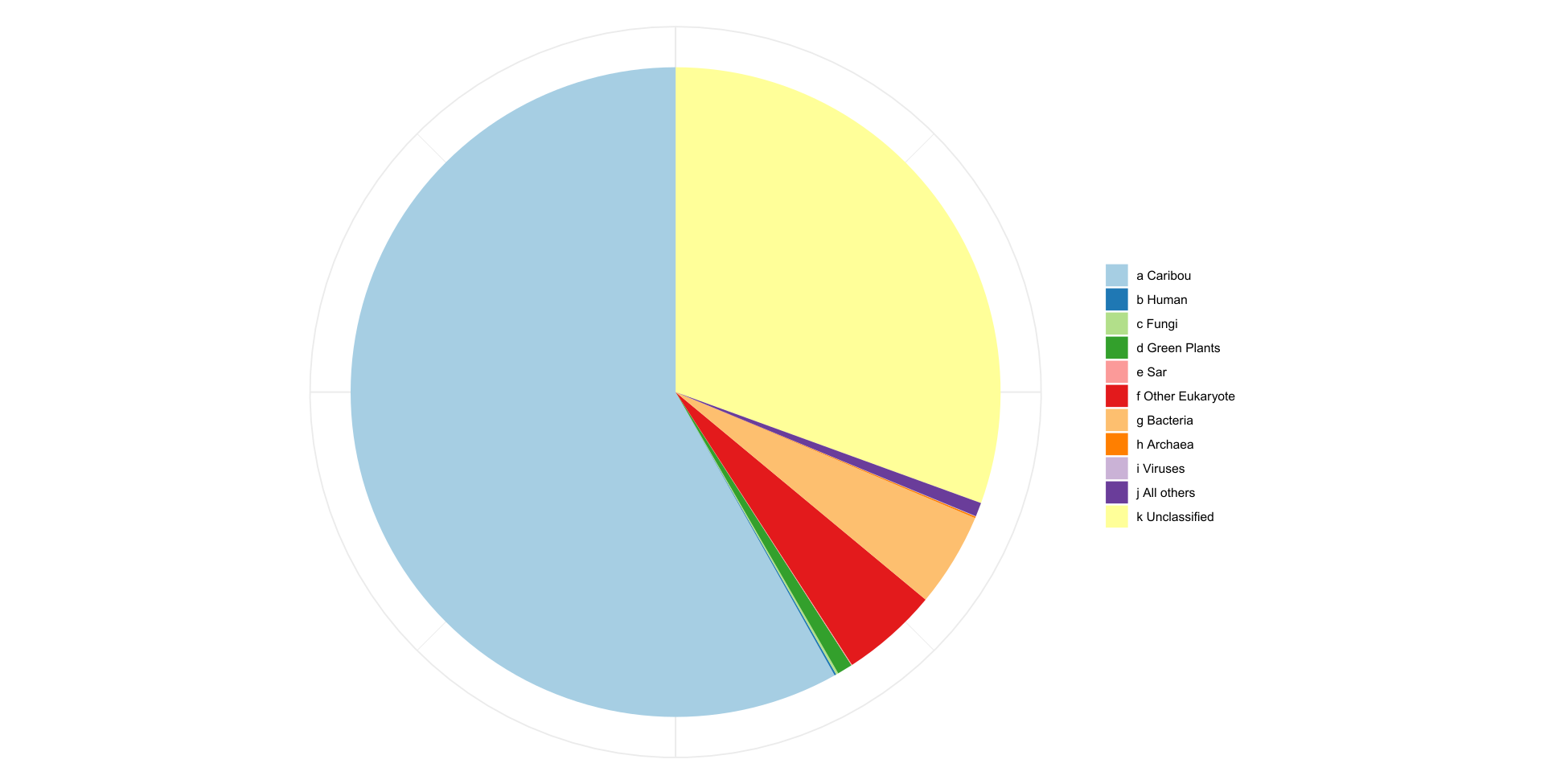
We are now successfully sequencing whole genomes from non invasively collected fecal samples, an important step towards the non invasive monitoring of caribou.

![]()